Science
Tokyo University Researchers Achieve Breakthrough in RNA Folding Simulations
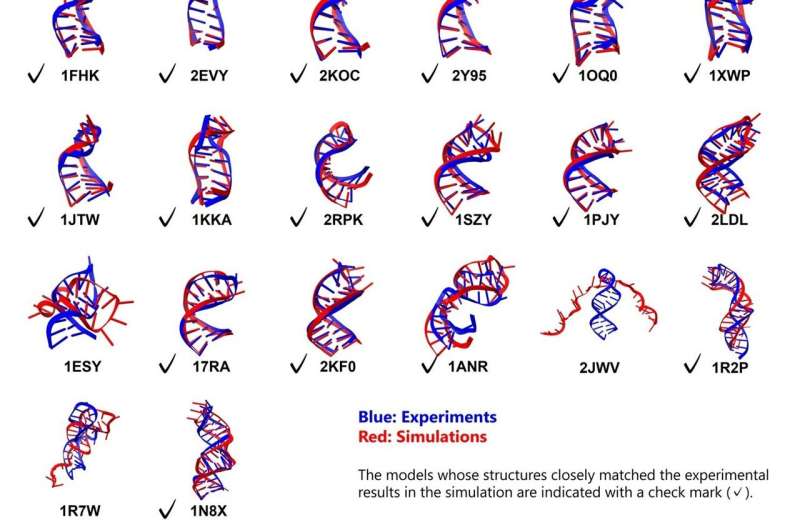
Researchers at Tokyo University of Science have made significant advancements in understanding RNA folding through new molecular dynamics simulations. Their findings, published on November 4, 2025, in the journal ACS Omega, demonstrate the ability to accurately simulate the folding process of RNA stem loops, which are crucial for various biological functions.
Ribonucleic acid (RNA) plays a vital role not only as a messenger for genetic information but also in processes such as gene regulation and maintenance across living organisms. The emergence of RNA-based therapeutics has heightened the need for precise modeling of its secondary and tertiary structures, which are critical for harnessing RNA’s full potential in biotechnology.
Advancements in Molecular Dynamics Simulations
Despite the importance of accurately simulating RNA folding, challenges have persisted in computational biophysics. Traditional methods often struggled to model the complete folding process from an unfolded state effectively. Most prior studies succeeded in accurately folding only a limited number of simple structures, primarily stem-loop formations.
In light of these challenges, Associate Professor Tadashi Ando from the Department of Applied Electronics at Tokyo University of Science conducted an extensive evaluation of modern simulation techniques. His research aimed to explore whether a combination of advanced computational tools could reliably model a diverse range of RNA stem loops.
The study utilized conventional molecular dynamics simulations, integrating two sophisticated components: the DESRES-RNA atomistic force field and the GB-neck2 implicit solvent model. The latter approximates the surrounding solvent as a continuous medium, significantly enhancing the speed of conformational sampling compared to traditional explicit solvent models, which are more computationally intensive.
Significant Findings and Future Implications
Dr. Ando’s research assessed a total of 26 RNA stem loops, with lengths ranging from 10 to 36 nucleotides, incorporating structures that featured bulges and internal loops. All simulations commenced from fully extended, unfolded conformations, leading to promising results. The study reported a high degree of folding accuracy, with 23 out of 26 RNA molecules achieving their expected structures. For the 18 simpler stem loops, the accuracy was particularly notable, with root mean square deviation (RMSD) values of less than 2 Å for the stems and less than 5 Å overall.
“Previous studies have reported only two or three folding examples of simple stem-loop structures, whereas this study addresses structures of much greater scale and complexity,”
Dr. Ando remarked.
Among the more complex motifs, five of the eight structures containing bulges or internal loops were accurately folded, revealing distinct folding pathways for these challenging configurations. This achievement marks a critical milestone in computational biology, validating the effectiveness of the combined force field and solvent models for studying large-scale conformational changes in RNA.
Although the study showcased high accuracy for stem regions, the loop regions demonstrated slightly lower precision, with RMSD values around 4 Å. This indicates the need for further optimization of the implicit solvent model, particularly in modeling non-canonical base pairs and accounting for the effects of divalent cations such as magnesium.
Overall, this research represents a pivotal advancement in the development of RNA-based medical tools. A deeper understanding of RNA folding is essential for designing drugs targeting RNA molecules, potentially leading to new treatments for genetic disorders, viral infections such as COVID-19 and influenza, and specific cancers.
Dr. Ando concluded, “The ability to reproduce the overall folding of the basic stem-loop structure is an important milestone in understanding and predicting the structure, dynamics, and function of RNA using highly accurate and reliable computational models. I expect this computational technique to lead to applications in RNA molecule design and drug discovery.”
As research continues to evolve, the implications of these findings may significantly impact the future of RNA-based therapies, paving the way for innovative medical solutions.
-

 Sports3 weeks ago
Sports3 weeks agoSteve Kerr Supports Jonathan Kuminga After Ejection in Preseason Game
-

 Top Stories2 weeks ago
Top Stories2 weeks agoMarc Buoniconti’s Legacy: 40 Years Later, Lives Transformed
-

 Science3 weeks ago
Science3 weeks agoChicago’s Viral ‘Rat Hole’ Likely Created by Squirrel, Study Reveals
-

 Politics3 weeks ago
Politics3 weeks agoDallin H. Oaks Assumes Leadership of Latter-day Saints Church
-

 Lifestyle3 weeks ago
Lifestyle3 weeks agoKelsea Ballerini Launches ‘Burn the Baggage’ Candle with Ranger Station
-

 Entertainment3 weeks ago
Entertainment3 weeks agoZoe Saldana Advocates for James Cameron’s Avatar Documentary
-

 Business3 weeks ago
Business3 weeks agoTyler Technologies Set to Reveal Q3 2025 Earnings on October 22
-

 Lifestyle3 weeks ago
Lifestyle3 weeks agoDua Lipa Celebrates Passing GCSE Spanish During World Tour
-

 Health3 weeks ago
Health3 weeks agoCommunity Unites for Seventh Annual Mental Health Awareness Walk
-

 Health3 weeks ago
Health3 weeks agoRichard Feldman Urges Ban on Menthol in Cigarettes and Vapes
-

 Business3 weeks ago
Business3 weeks agoMLB Qualifying Offer Jumps to $22.02 Million for 2024
-

 Sports3 weeks ago
Sports3 weeks agoPatriots Dominate Picks as Raiders Fall in Season Opener









